To streamline the cybershake workflow it would be good to include source generation and realisation installation as tasks that can be completed by the workflow.
Current status
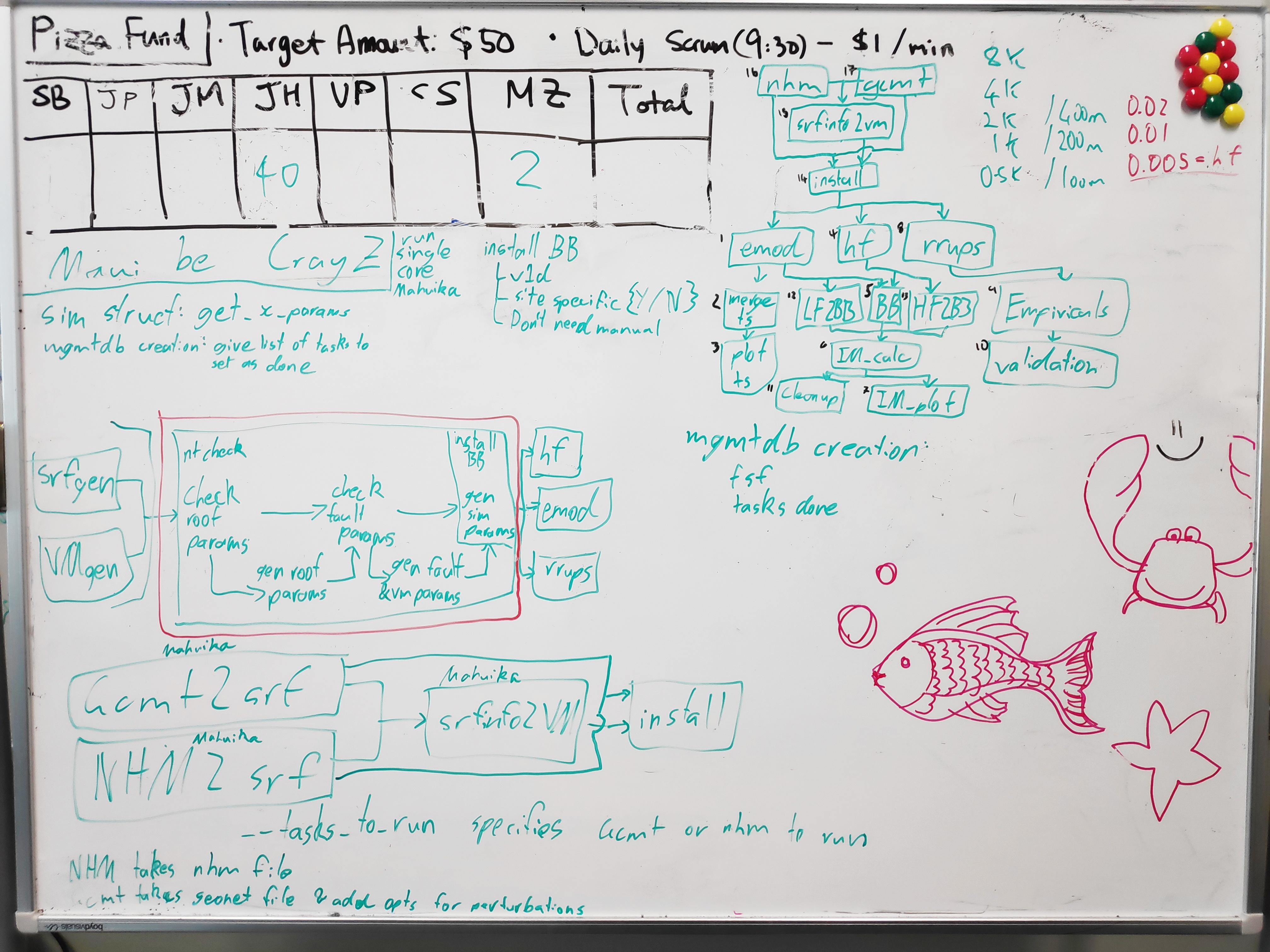
Currently to prepare sources and get them ready for a cybershake the following steps must be taken:
- Generate srfs using either GCMT2SRF or NHM2SRF with a list of all faults and realisation counts
- Generate velocity models for each fault using srfinfo2vm (This can be done one fault at a time, but is designed to do all at once)
- Install each realisation using the install_cybershake script. This also prepares the database and queue management folder
As each of these steps is designed to run on an entire fault or cybershake simulation at once, we are not able to directly integrate them into the workflow
Proposed changes
In order to integrate each of these processes into the automated workflow, they must be refactored to run on a single realisation at a time.
For gcmt2srf and nhm2srf this presents no major issues aside from the refactor itself.
Neither of these tasks should be set as the default as it is not trivial to determine which process should be run. This is further complicated if there are additional configuration options required for a run.
This means that without one being explicitly defined nothing will run and the automated wrapper/auto submit scripts will terminate very quickly.
As each realisation in a fault uses the same velocity model it is important that only one realisation of each fault generates the velocity model.
This can be set in the configuration file of the run_cybershake script, meaning that only the first realisation in each fault runs the task.
In order to allow all processes to know that the vm has been created the add_to_queue script can be called with "<fault_name>_REL%" as the realisation on successful completion of the srfinfo2vm script in order to update the velocity model generation task for all realisations of the fault in the database.
The dependencies for srfinfo2vm are one of either gmct2srf or nhm2srf, but without a default.
Installation of each realisation will need to check for the existence of root_params and fault_params before attempting to generate them as the first installation task run will need to be responsible for generating root_params, and the first installation task run of each fault will be responsible for generating fault_params and updating vm_params.
This could be done by a file existence check, where the first task to not find each shared file creates a dummy file while it generates the contents, and each other task waits for the file to be completed before moving to the next step. Alternatively dependencies could be changed such that all tasks after the first of a realisation are dependent on the first task completing.
Install_BB is able to be refactored into the main root_params generation now.
Installation will no longer be responsible for generation of the database.
As the database will be required for the source generation and installation steps the database should be created before run_cybershake is called.
The database creation script should only require the location of the cybershake root directory and a fault selection file, as given in Step 2 of the cybershake manual.
Allowing a race condition on installation between faults trying to generate root_params is acceptable, provided the final product is the full file. Likewise for realisations generating fault_params.
srfinfo2vm should still only be done once per fault however.
- This can run with the ONCE keyword by default. The slurm script can use "<fault_name>_REL%" to update all other tasks as previously noted.
add_to_queue will need to have a flag added to allow for the update within the file to use a wildcard realisation instead of the number for that realisation. The name of te file should
The expected workflow dependency tree after this refactor has occurred is available here: Workflow dependencies
Draft changes
A record of the whiteboard with the draft changes for this refactor are available below: